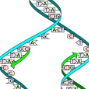
To begin with, DNA has no amino acids, which are found in proteins. Nucleotide bases hold the DNA strands together through hydrogen bonding.
Until I see the sequence and the primers used I do not believe Mayor's claim of chimpanzee DNA. Based on behavior and physical characteristics it's as unlikely that Sasquatch DNA is that close to a chimpanzee as it is to be very close to human. UNLESS hybridization is involved. Presumably we are talking about mitochondrial DNA, which is inherited from the mother only. So a hybridization event will not show anything about evolution of the male, or the female. It's a slice in time(of the mating) of the female. Nuclear eDNA is much more difficult (and expensive) to sequence, and was not likely the case in either the Mayor or the Disotell case. But, please, show me the data. If Mayor used a sequencer that had previously been used on chimpanzee DNA, there could be carry over. Protocol details, especially blanks and standards, would be helpful to know here.
Comparisons of the Kentucky vs the Washington environment based only on rainfall neglect other important factors such as microbe species and populations, temperatures, and sample handling. There is plenty of opportunity for degradation.
I am currently analyzing littoral eDNA sequences for signs of an unknown primate and have learned that sequencing errors can confuse the issue, as well as heteroplasmy, and the possibility of sperm mtDNA leaking (into the egg). The latter is fairly minute in humans but may not be so in Sasquatch.
The community awaits a sample collected from an observed Sasquatch immediately after deposition, or a body part. Otherwise, as mentioned above, there are too many unknowns to base a case on subtle differences. In the mtDNA region of over 200 bases that I studied, Neanderthal differs from modern human by only ONE mutation, so there's "no room" to distinguish an intermediate Sasquatch there. Longer sequences in other regions are desirable.
There's a lot of data to sort through in this work. The so called "mammalian" primers I used also sequenced birds, and fish, lots of them. Unfortunately I know of no readily available software to do this. Also, the NCBI BLAST results are not eDNA friendly, so relevant data must be extracted through character manipulation of large flat files. I wrote BASIC programs and also used Excel sorting. A goal of this work is to develop a simple procedure that can be used by our Community to analyze sequence data from commercial labs.